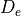Computing energy levels¶
In the following we present all keywords and options relevant to the calculations of energy levels.
Calculation (input) setup¶
Atoms: defines the chemical symbols of the two atoms.
Example:
atoms Na-23 H-2
specifies the 23NaD diatomic. Duo includes an extensive database of atomic properties (atomic masses,
nuclear spins, isotopic abundances and other quantities) and will use the appropriate values
when required. The database should cover all naturally-occurring nuclei as well as all radioactive ones
with a half-life greater than one day and is based on the AME2012 and
NUBASE2012 databases. Each atom should be specified by its chemical symbol,
a hyphen (minus sign) and the atomic mass number, like in the example above.
Atomic masses will be used, which is generally the most appropriate choice unless one is explicitely including
non-adiabatic corrections. The hydrogen isotopes deuterium and tritium can also
be optionally specified by the symbols D and T.
The atomic mass number can be omitted, like in the following example:
atoms Li F
In this case Duo will use the most-abundant isotopes (7Li and 19F in the example above) or, for radioactive nuclei not naturally found, the longest lived one. For example
atoms Tc H
selects for technetium the isotope 97Tc, which is the longest-lived one. A few nuclides in the database are nuclear metastable isomers, i.e. long-lived excited states of nuclei; these can be specified with a notation of the kind
atoms Sb-120m H
In the example above the radioactive isotope of antimony 120mSb is specified (and hydrogen). Another example
atoms Sc-44m3 H
specifies the scandium radioactive isotope 44m3Sc (and hydrogen).
masses:
This is an optional keyword which specifies explicitely the masses of the two atoms (in Daltons, i.e. unified atomic mass units), overriding the values from the internal database if the keyword texttt{atoms} is also specified.
For example, the masses for the CaO molecule would be:
masses 39.9625906 15.99491463
The masses may be atomic masses (the recommended choice if one does not include adiabatic or non-adiabatic corrections), nuclear masses. An up-to-date reference of atomic masses is provided by the AME2012 catalogue (Chin. Phys. C, 36:1603–2014, 2012.) Duo can also make use of position-dependent masses (which is a practical way to account for non-adiabatic effects), see Bob-Rot.
nstates: is the number of potential energy curves (PECs) included in the calculation.
For example, if the ground state and four excited states of a molecule are to be included:
nstates 5
Note that if nstates is set to a number different from the actual number of PECs included in the
input file no error message is issued; if more than nstates PECs are included in the input file then the PECs
with state > nstates will be ignored.
Note also that, consistently with the way Duo works internally, nstates is the number of unique PECs in absence of spin-orbit couplings.
jrot: specifies the set of total angular momentum quantum numbers to be computed.
These must be integers or half-integers, depending on whether there is an even or odd number of electrons. One can directly specify the values (separated by spaces or commas), specify a range of values (a minimum and a maximum values separated by a hyphen; note than the hyphen must be surrounded by at least by one space on each side). The values do not have to appear in ascending order. For example, the following line
jrot 2.5, 0.5, 10.5 - 12.5, 20.5
specifies the set J = 0.5, 2.5, 10.5, 11.5, 12.5, 20.5.
The first J in the jrot list will be used to define the reference zero-point-energy (ZPE) value for the
run.
Note that in the optional sections specifying calculation of spectra (See Intensity) or
specifying fitting (section ref{sec:fitting}) is necessary to specify again a list of
J values by J and Jlist respectively, which are completely independent from the jrot value
specified for energy level calculation.
symmetry: (or
Symgroup) is an optional keywork specifying the molecular permutation-inversion symmetry group.
Cs(M) is for heteronuclear diatomics and C2v(M) is for homonuclear diatomics.
For example:
symmetry Cs(M)
Instead of C2v(M) one can write equivalently C2h(M) or G4(M), as these groups are isomorphic;
the only difference will be in the labels used for the energy levels.
The short-hand notations Cs, C2v, C2h and G4 can also be used and are equivalent to the ones with (M).
The energy calculations are done using Cs(M), which is also the default, while for the intensities the C2v(M) group can be also used.
Note that this keyword refers to the symmetry of the exact total (electronic, vibrational and rotational)
Hamiltonian and not to the 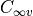 or the
or the 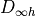 point groups, which are relative
to the clamped-nuclei electronic Hamiltonian.
point groups, which are relative
to the clamped-nuclei electronic Hamiltonian.
DO_NOT_SHIFT_PECS: suppresses shifting the PEC to the minimum of potential 1 (assumed to the lowest).
The default is to do the shift of the PECs to the minimum of poten 1. In order to suppress shifting energies to ZPE, use
ZPE 0.0
see also the description of the keyword ZPE.
SOLUTIONMETHOD * defines the DVR basis set and thus the DVR solution method for the vibrational problem.
Possible methods include 5POINTDIFFERENCES.
SOLUTIONMETHOD 5POINTDIFFERENCES
for the 5 points stencil finite differences to derive the kinetic energy operator. A more efficient method is Sinc DVR (default), which is switched on with
SOLUTIONMETHOD SINC
Since Sinc is also currently the default method, this does not have to be specified.
Defining the grid¶
grid: specifies an input section with the specifications of the grid of points.
It is used for the solution of the vibrational problem.
Example:
grid
npoints 501
unit angstroms
range 1.48 , 2.65
type 0
end
Keywords
npoints: is the number of grid points Np.
Typical runs use 100 to 500 points.
npoints 301
- units: is optional and specifies the unit of measure of the grid specifications; possible values are
angstroms(default) or
bohrs.
- units: is optional and specifies the unit of measure of the grid specifications; possible values are
range: specifies the range of the grid in terms of rmin and rmax the lower and upper bond lengths.
rmin should be strictly greater than zero and rmax strictly greater than rmin. As elsewhere in the program, the value may be separated by a space or a comma.
type: Grid type
Is an integer number 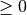 which specifies the type of grid.
which specifies the type of grid.
Duo support not only uniformely spaced grids (default), which correspond to type 0,
but also various kind on non-uniformely spaced ones, which are particularly useful for near-dissociation,
very weakly bound states.
Example:
type 0
In the case of uniformely-spaced grids the mesh points rj, j=0, Np-1 are given by
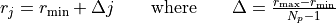
Non-uniformely spaced grids are based on a change of variables from r to z=f(r); it is then
the transformed variable z that is uniformely sampled. The transformed variables z are
parametrised by two parameters, 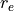 and
and 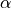 , which have to be specified
for the grid types > 0 (see below).
, which have to be specified
for the grid types > 0 (see below).
Transformed variable currently implemented are (Phys. Rev. A, 78:052510, 2008):
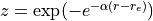
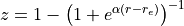
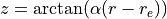
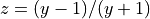 with
with 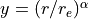
All the transformed grids have the property of decreasing the density of points for large r, so that one does not waste too many points in regions where the potential is almost constant and the corresponding vibrational wave function slowly varying.
re: (alias
ref) Reference bond length used fortype> 0 (see above).alpha: Parameter
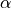 for
for type >0(see above).
Eigensolver¶
The input section EigenSolver (aliases: FinalStates, diagonalizer, FinalStates)
specifies
various options relative to the J>0 and/or the coupled problem; it also specifies
the LAPACK routine which should be used for matrix diagonalization (both for the solution of the
vibrational problem and for the solution of the coupled problem).
Example:
Eigensolver
enermax 25000.0
nroots 500
ZPE 1200.0
SYEVR
END
Keywords
nroots is the number of energy levels of the coupled problem to be computed (for any of the specified values of
jrot).
Example:
nroots 500
enermax: is an energy threshold
Enermax (aliases: uplimit, enercut) is to select the energy levels of the coupled problem to be computed (cm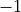 ).
).
For example:
enermax 15000.
If both nroots and enermax are specified then only levels satisfying both criteria are selected.
Note that the present enermax threshold
is distinct from the homonymous one in the vibrationalbasis input section, as the latter refers to the solution of the J=0
uncoupled problem while the one being discussed
at present refers to the solution of the full (rotationally excited and/or coupled) problem.
ZPE: Zero point energy
ZPE allows to explicitly input the zero-point energy (ZPE) of the molecule (in cm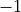 ). This affects the value printed, as
Duo always prints energy of rovibronic levels by subtracting the ZPE. Example:
). This affects the value printed, as
Duo always prints energy of rovibronic levels by subtracting the ZPE. Example:
ZPE 931.418890
If ZPE is not included Duo will define the ZPE value as the lowest computed energy for the first value of J
listed next to the jrot keyword (jlist), from the positive parity block. Currently it is not possible to
take an automatic ZPE from the negative parity block (it is however possible in the intensity and fitting parts of the output).
Thus ZPE does not necessarily have to be from the ground electronic state.
This ZPE taken from the eigensolver/diagonalizer section changes the energies in the main, standard Duo output.
The ZPE shift can be suppressed by setting the ZPE value to zero. This should be done either in the Diagonalizer, Fitting or Intensity
sections, depending on the current task:
ZPE 0.0
SYEVR or SYEV: LAPACK Eigesolvers
This optional keywords permits to specify which routine from the LAPACK library should be used for matrix diagonalization. At the moment only the two options quoted are implemented. Example:
SYEV
The SYEV routine (default) first reduces the matrix to diagonalize to tridiagonal form using orthogonal similarity transformations, and then the QR algorithm is applied to the tridiagonal matrix to compute the eigenvalues and the eigenvectors. The SYEVR routine also reduces the matrix to diagonalize to tridiagonal form using orthogonal similarity transformations but then, whenever possible, computes the eigenspectrum using Multiple Relatively Robust Representations (MR). SYEVR might give better performance, although exact timings are system- and case-dependent.
ASSIGN_V_BY_COUNT
The vibrational quantum number  is assigned by counting the rovibronic states of the same
is assigned by counting the rovibronic states of the same State, 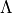 ,
,
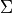 arranged by increasing energy. The corresponding
arranged by increasing energy. The corresponding State, 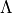 ,
,
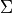 labels are defined using the largest-contribution approach
(the quantum labels corresponding to the basis set contribution with the largest expansion coefficient).
The keyword should appear anywhere in the body of the input file. The default is to use the largest-contribution
approach also to assign the vibrational quantum number (no
labels are defined using the largest-contribution approach
(the quantum labels corresponding to the basis set contribution with the largest expansion coefficient).
The keyword should appear anywhere in the body of the input file. The default is to use the largest-contribution
approach also to assign the vibrational quantum number (no ASSIGN_V_BY_COUNT).
Example: computing energy levels (one PEC)¶
Here below there is a commented, minimalistic Duo input file for a single Morse potential; note that the input is case-insensitive.
In this particular example we compute the 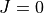 energy levels of a Morse oscillator
energy levels of a Morse oscillator 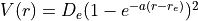 with
with 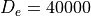 cm-1,
cm-1,
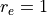 Angstrom and
Angstrom and 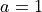 Angstrom
Angstrom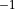 ; the masses of both atoms are both set to 1 Dalton, so that this example
is very approximately corresponds to the hydrogen molecule H:math:_2. The exact energy levels are given by
; the masses of both atoms are both set to 1 Dalton, so that this example
is very approximately corresponds to the hydrogen molecule H:math:_2. The exact energy levels are given by
![E_n = \omega (n+1/2) \left[1 - x_e (n+1/2) \right]](_images/math/943ec12833bbad3e913d8288b6c3683e2bf8d399.png) ,
, 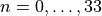 ,
with
,
with 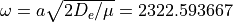 cm-1 and
cm-1 and 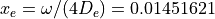 .
.
(DUO test input)
masses 1.00000 1.000000
nstates 1
jrot 0 10
grid
npoints 250
range 0.30, 6.50
end
EigenSolver
enermax 35000.0
nroots 10
SYEV
end
VibrationalBasis
vmax 10
END
poten 1
name "Morse"
type Morse
lambda 0
mult 1
symmetry +
units cm-1
units angstroms
values
v0 0.000000
r0 1.000000
a0 1.000000
De 40000.
end
The output has this structure:
Input line |
Description |
|---|---|
(DUO test input) |
comment line |
masses 1.00000 1.000000 |
masses of the two atoms, in Daltons** |
nstates 1 |
number of PECs in the input |
jrot 0 10 |
total angular momentum J |
grid |
specification of the grid |
npoints 250 |
number of grid points |
range 0.30, 6.50 |
|
end |
end of grid specification |
EigenSolver |
options for the Eigensolver |
enermax 35000.0 |
print only levels up to enermax cm-1 |
nroots 10 |
print only nroots lowest-energy levels |
SYEV |
use SYEV diagonalizer from LAPACK |
end |
end of input section EigenSolver |
VibrationalBasis |
options for the vibrational uncoupled problem |
vmax 10 |
compute vmax}+1 vibrational states |
end |
end of vibrational specifications |
poten 1 |
PEC number 1 specification |
name “Morse” |
label |
type Morse |
functional form: (extended) Morse function |
lambda 0 |
quantum number |
mult 1 |
multiplicity, |
symmetry + |
only for |
units cm-1 |
unit for energies |
units angstroms |
unit for distances and inverse distances |
values |
beginning of specification of the parameters |
v0 0.000000 |
specification of global shift |
r0 1.000000 |
specification of |
a0 1.000000 |
specification of |
De 40000. |
specification of |
end |
end of PEC number 1 specification |
Duo will by default echo the whole of the input file in the output between the lines (Transcript of the input --->) and
(<--- End of the input). This is useful so that the ouput file will also contain the corresponding input.
To avoid echoing the input just add the keyword do_not_echo_input anywhere in the input file (but not within an input section).
Duo will then print its logo, the values of the physical constants (used by the program for such things as conversions between different units) and print some of the global input parameters such as the number of grid points, extent of the grid etc.
- Duo will then print the values of all objects (PECs, dipole moment curves, couplings) on the internal grid. For PECs Duo will also
compute and print quantities such as the value of the first few derivatives at the minimum, the corresponding equilibrium spectroscopic constants (harmonic frequency, rigid-rotor rotational constant etc.).
Duo will solve the
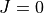 one-dimensional Schroedinger equation for each of the PECs and print the corresponding
one-dimensional Schroedinger equation for each of the PECs and print the corresponding
vibrational (contracted)energies.Duo will then solve the full problem (with
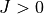 and/or all coupling terms activated).
In the example above we specified two values of
and/or all coupling terms activated).
In the example above we specified two values of 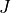 , namely
, namely 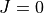 and
and 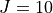 . The
. The  energies will be exactly the same as the
energies will be exactly the same as the vibrational (contracted)ones, as in our example there are no couplings at all.
The Duo input files for this example can be found in [Duo Tutorial](https://github.com/Trovemaster/Duo/tree/MOLPRO/examples/tutorial)
See [The ab initio ground-state potential energy function of beryllium monohydride, BeH by Jacek Koput, JCP 135, 244308 (2011)](http://dx.doi.org/10.1063/1.3671610)
The ground electronic state of BeH is a doublet (2Sigma+), see [https://www.ucl.ac.uk/~ucapsy0/diatomics.html](https://www.ucl.ac.uk/~ucapsy0/diatomics.html).
Example: BeH in its ground electronic state¶
In order to solve the nuclear motion Schroediner equation to compute ro-vibronic spectra of BeH with Duo we need to prepare an input file using the following structure (BeH_Koput_01.inp):
atoms Be H
(Total number of states taken into account)
nstates 16
(Total angular momentum quantum - a value or an interval)
jrot 0.5 - 2.5
(Defining the integration grid)
grid
npoints 501
range 0.4 8.0
type 0
end
CONTRACTION
vib
vmax 30
END
poten 1
units cm-1 angstroms
name 'X2Sigma+'
lambda 0
symmetry +
mult 2
type grid
values
0.60 105169.63
0.65 77543.34
0.70 55670.88
0.75 38357.64
0.80 24675.42
0.85 13896.77
0.90 5447.96
0.95 -1125.87
1.00 -6186.94
1.05 -10024.96
1.10 -12872.63
1.15 -14917.62
1.20 -16311.92
1.25 -17179.13
1.30 -17620.16
1.32 -17696.29
1.33 -17715.26
1.34 -17722.22
1.35 -17717.69
1.36 -17702.19
1.37 -17676.19
1.38 -17640.16
1.40 -17539.76
1.45 -17142.53
1.50 -16572.59
1.55 -15868.72
1.60 -15063.34
1.65 -14183.71
1.70 -13252.86
1.80 -11313.
1.90 -9369.74
2.00 -7518.32
2.10 -5832.29
2.20 -4366.71
2.30 -3155.94
2.40 -2208.98
2.50 -1507.72
2.60 -1013.23
2.80 -456.87
3.00 -221.85
3.50 -72.13
4.00 -41.65
4.50 -24.9
5.00 -14.32
6.00 -4.74
8.00 -0.75
10.00 -0.19
20.00 0.0
end
where we use the potential energy curve (PEC) defined in Table III of Koput J. Chem. Phys. 135, 244308 (2011) in a grid form.
An alternative definition is an analytical PEC, see e.g. Barton et. al MNRAS 434, 1469 (2013)
poten 1
units cm-1 angstroms
name 'X2Sigma+'
lambda 0
symmetry +
mult 2
type grid
values
V0 0.00
RE 1.342394
DE 17590.00
RREF -1.00000000
PL 3.00000000
PR 3.00000000
NL 0.00000000
NR 0.00000000
b0 1.8400002
end
 and
and  , in Angstroms
, in Angstroms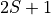
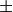 symmetry
symmetry